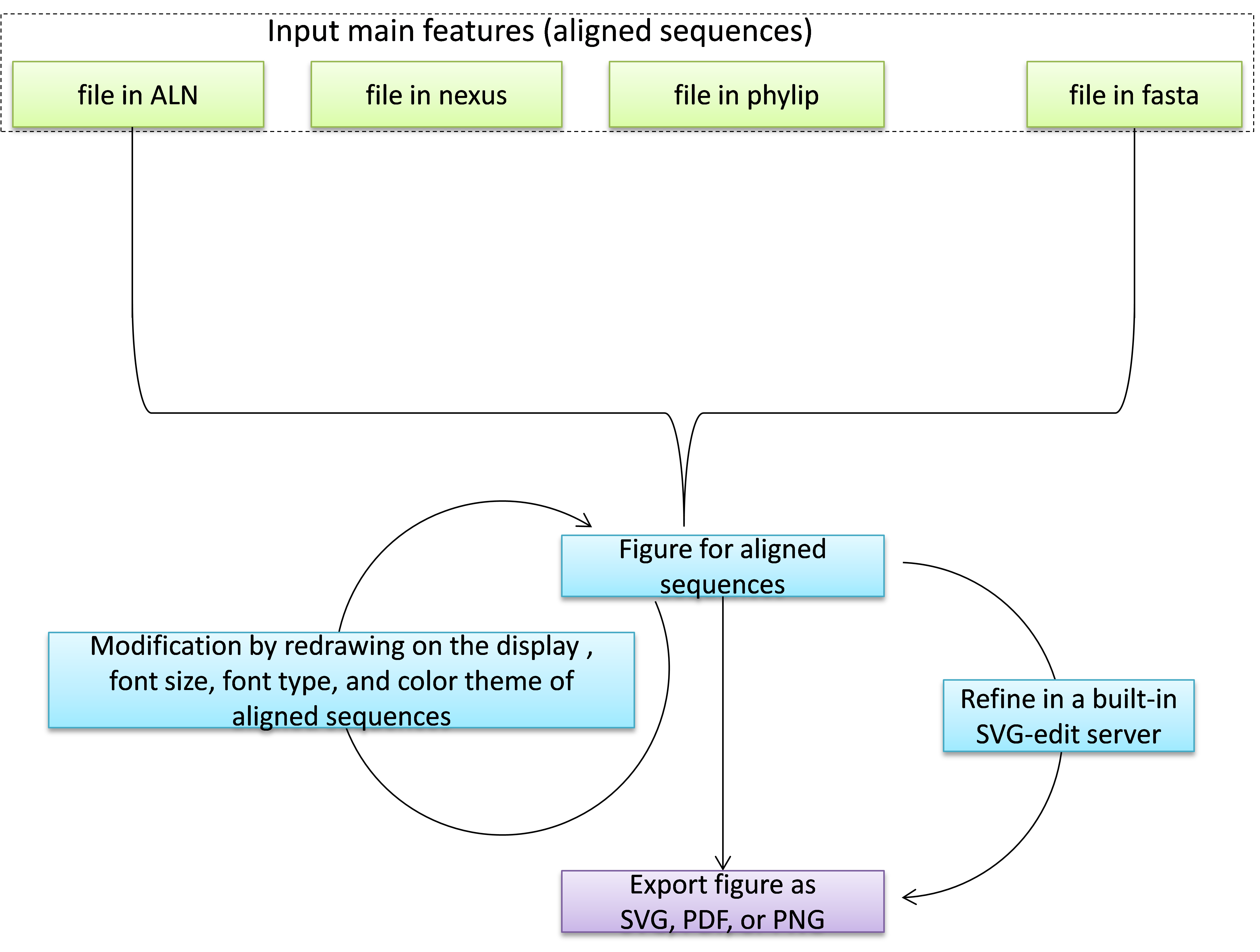
aln2svg is designed for the visualization of aligned sequences, and the generation of high-quality figures for publication. aln2svg supports features in four types of formats, including aln, nexus, phylip, and fasta. After inputting these features, aln2svg would transform them into a svg format.
 Download high-resolution work flow in PowerPoint
Download high-resolution work flow in PowerPoint
After the first generation of figures, Users can customize the font size, font type, sequence length per line and color of sequences after their initial render and even fine-tune each element through an integrated visual editor. Finally, the generated figure can be exported as either vector graphic (in SVG and PDF format), or raster graphic (in PNG format).
Users can input the sequences in four formats, including aln, nexus, phylip, and FASTA Sequences. Brief introduction for these formats are described as following:
This format is used by many popular programs like CLUSTAL W, Clustal X, T-Coffee, MAFFT, MUSCLE. CLUSTAL file names often have a .aln extension.
Example:
CLUSTAL format alignment by MAFFT (v7.475)
DL328 atggcaaagatgtcatcttcagatcctttagttcttggaagagtggttggagatgtcatc
KOP atggcaaagatgtcatcttcagatcctttagttcttggaagagtggttggagatgtcatc
shitouqi atggcaaagatgtcatcttcagatcctttagttcttggaagagtggttggagatgtcatc
haihua atggcaaagatgtcatcttcagatcctttagttcttggaagagtggttggagatgtcatc
BBG51B.1 atggcaaagatgtcatcttcagatcctttagttcttggaagagtggttggagatgtcatc
************************************************************
DL328 cactctttcaatccaagtgttcaaatgtctgtcacttacaacaccaaacaagtcttcaat
KOP cactctttcaatccaagtgttcaaatgtctgtcacttacaacaccaaacaagtcttcaat
shitouqi cactctttcaatccaagtgttcaaatgtctgtcacttacaacaccaaacaagtcttcaat
haihua cactctttcaatccaagtgttcaaatgtctgtcacttacaacaccaaacaagtcttcaat
BBG51B.1 cactctttcaatccaagtgttcaaatgtctgtcacttacaacaccaaacaagtcttcaat
************************************************************
Note: the sequences must be in same length.
This is the file format used by many popular programs like GARLI, GDA, MacClade, Mesquite, ModelTest, MrBayes and PAUP*. Nexus file names often have a .nxs or .nex extension.
Example for nexus (see more details)
#NEXUS
BEGIN DATA;
dimensions ntax=4 nchar=464;
format missing=?
interleave datatype=PROTEIN gap= -;
matrix
AH01G000010. M------------------------------------------------G
AH01G000020. MSVAADSPIHSSSSDDFIAYLDDALAASSPDASSDKEVENQDELESGRIK
AH01G000030. --------------------------------------------------
AH01G000040. MRYL------------IIFFLPSCY------------IMEEDKLPMG--S
AH01G000010. KC------------------QAMHKYV----GVF----------------
AH01G000020. RCKFESAEETEESTSEGIVKQNLEEYVCTHPGSFGDMCIRCGQKLDGESG
AH01G000030. --------------------------------------------------
AH01G000040. DCSFYSFE----------------------------LC----------SG
AH01G000010. ---------------------------------LALLVAFNHTHLTQGRK
AH01G000020. VTFGYIHKGLRLHDEEISRLRNTDVKNLLIRKKLYLILDLDHTLLNSTH-
AH01G000030. --------------------------------------------------
AH01G000040. RHFGTTY-------------------------------------------
Note: the sequences must be in same length.
The PHYLIP format came from Joe Felsenstein’s phylogeny inference package and is now used by several phylogenetics programs. PHYLIP file names often have have a .phy or .ph extension.
Example (see more details):
4 464
AH01G001 M--------- ---------- ---------- ---------- ---------G
AH01G002 MSVAADSPIH SSSSDDFIAY LDDALAASSP DASSDKEVEN QDELESGRIK
AH01G003 ---------- ---------- ---------- ---------- ----------
AH01G004 MRYL------ ------IIFF LPSCY----- -------IME EDKLPMG--S
KC-------- ---------- QAMHKYV--- -GVF------ ----------
RCKFESAEET EESTSEGIVK QNLEEYVCTH PGSFGDMCIR CGQKLDGESG
---------- ---------- ---------- ---------- ----------
DCSFYSFE-- ---------- ---------- ------LC-- --------SG
---------- ---------- ---------- ---LALLVAF NHTHLTQGRK
VTFGYIHKGL RLHDEEISRL RNTDVKNLLI RKKLYLILDL DHTLLNSTH-
---------- ---------- ---------- ---------- ----------
RHFGTTY--- ---------- ---------- ---------- ----------
Note: the sequences must be in same length.
Example for FASTA sequences (see more details):
>AH01G000010. M------------------------------------------------GKC-------- ----------QAMHKYV----GVF------------------------------------ -------------LALLVAFNHTHLTQGRKIIKPLNTNNINPHQPITLDHDSSSVGDSGK KSMVV---LLESLEAKKVSVNGVGPGPSPGAGHR-------------------------- ---------------KTDGKDN-VMKSMGAVVV---VVKNNQDVK---DS---------- ------------------------------------------------------------ ---------------------------INGVGP---------GHS------PGVGH---- ------------------------------------------KN >AH01G000020. MSVAADSPIHSSSSDDFIAYLDDALAASSPDASSDKEVENQDELESGRIKRCKFESAEET EESTSEGIVKQNLEEYVCTHPGSFGDMCIRCGQKLDGESGVTFGYIHKGLRLHDEEISRL RNTDVKNLLIRKKLYLILDLDHTLLNSTH--LAHLNSEELH------LISQADSLGDVSK GSLFKLDKMHMMTKLRPFVRTFLKEASEM---FEMYIYTMGDRPYALE------------ MAKLLDPLGEYFNA-KVISRDDGTQKHQKGLDI---VLGQESAVVILDDTEHAWVKHKDN LILMERYHFFGSSCRQFGFNCKSLAELKSDEDEAEGALTKILKVLKQVHSKFFDELKEDI AERDVRQVLKSVRREVLSGCVVVFSRIFHGALPPLRQMAEQLGATCLMELDPSVTHVVAT DAGTEKARWAVKEKKFLVHPRWIEAANYFWEKQPEENFVLKKKQ
Note: the sequences must be in same length.
Example for result page:

After the first generation of the figure, users can modify it by setting on the "region 2" in the result page, including modifying the color, sequence length per line, font type, and font size of the features.
Besides that, users can also send the generated figure to the built-in SVG-editor for further refining by clicking "Edit figure interactively".
Finally, users can save the figure as SVG, PNG, or PDF.
Users can download the source code of aln2svg here, and install it locally.